This protocol can be adapted to use dUTP or dCTP labelled with other dyes, biotin, or haptens like digoxigenin. See Biotium’s full selection of labelled nucleotides.
| Materials required: | Workflow overview: |
|
|
* When using dUTP conjugates for labelling, use Taq DNA polymerase; dUTP inhibits archaeal polymerases such as Pfu and Vent®.
Procedure:
1 – Set up labelling reactions
1.1 For each labelling reaction, set up the PCR reaction mix as shown below:
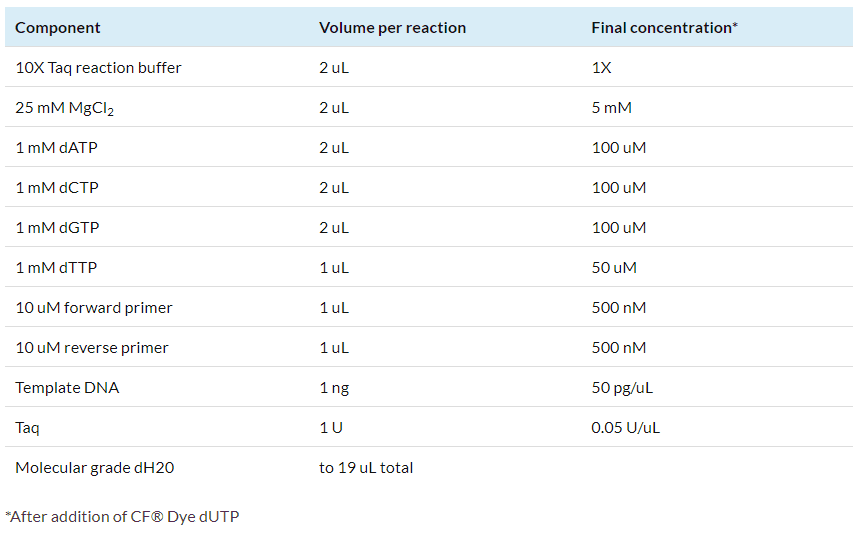
1.2 Add 1 uL of 1 mM CF® dye dUTP to the reaction tube.
-
- Optional: for an unlabeled control reaction, add 1 uL of 1 mM dTTP instead of CF® dye dUTP.
- If using fluorescent dCTP, set up the reaction with 50 uM dCTP and 100 uM dTTP, then add 1 uL of 1 mM CF® Dye dCTP to the reaction.
2 – Perform PCR amplification
Amplify the reactions in a thermocycler using the following cycling protocol:
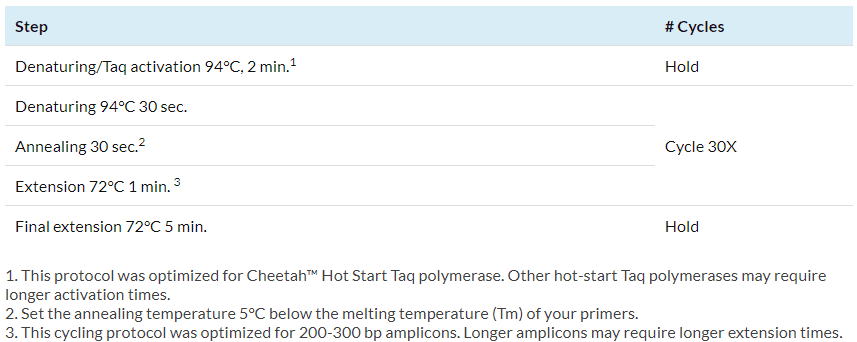
3 -Remove unincorporated nucleotides
Use a PCR clean-up kit or G50 Sephadex® microspin column to remove unincorporated nucleotides.
- Removal of unincorporated nucleotides may not be necessary before hybridization, but the fluorescence from free labelled nucleotides can make it difficult to evaluate labelled PCR products by gel electrophoresis.
4 – Evaluate labelling by gel electrophoresis
4-1 Run 10% of the labelled product on an agarose gel along with a DNA ladder. Do not add fluorescent DNA dye to the agarose before casting. After electrophoresis, image the CF® dye fluorescence of the labelled probes on a UV gel transilluminator or laser-based gel scanner as appropriate for the wavelengths of the specific dye used.
- It can be useful to run an unstained DNA ladder in one lane, and a DNA ladder prestained with GelRed® Prestain Plus 6X DNA Loading Buffer in another lane to visualize the ladder before staining the entire gel.
- Visible fluorescent dyes (CF®405S to CF®594) can be viewed with UV excitation. Far-red fluorescence emission (650 nm or longer) is not visible to the human eye, but can be imaged using a fluorescence gel scanner using the appropriate excitation and emission settings.
- Be sure to image CF® dye fluorescence before staining DNA with gel stain, because CF® dye fluorescence may overlap with gel stain fluorescence, or CF® dyes and gel stains may quench one another.
4-2 After imaging the probe fluorescence, post-stain the gel with a nucleic acid gel stain like GelRed® or GelGreen® to image unstained DNA ladder and unlabelled control PCR product.
- Fluorescent dyes may cause shifts in DNA migration of the labelled DNA compared to unlabelled PCR product.
 |
Water, Ultrapure Molecular Biology Grade |
 |
Cheetah™ PEA-PLEX HotStart Taq DNA Polymerase |
 |
dNTP Set, 100 mM Each |
 |
dUTP CF® Dye Conjugates |
 |
dCTP CF® Dye Conjugates |
